The parameters are reduced parameters. More...
Namespaces | |
| AntiochPrivate | |
| Constants | |
| KineticsModel | |
| ReactionType | |
| UnitBaseConstant | |
| UnitBaseStorage | |
Typedefs | |
| typedef unsigned int | Species |
| typedef DefaultSourceFilename | DefaultFilename |
Enumerations | |
| enum | ParsingType { ASCII = 0, XML, CHEMKIN } |
| enum | ParsingKey { SPECIES_SET = 0, SPECIES_DATA, SPECIES, THERMO, PHASE_BLOCK, TMIN, TMAX, NASADATA, NASA7, NASA9, REACTION_DATA, REACTION, REVERSIBLE, ID, EQUATION, CHEMICAL_PROCESS, KINETICS_MODEL, REACTANTS, PRODUCTS, FORWARD_ORDER, BACKWARD_ORDER, PREEXP, POWER, ACTIVATION_ENERGY, BERTHELOT_COEFFICIENT, TREF, HV_LAMBDA, HV_CROSS_SECTION, UNIT, EFFICIENCY, FALLOFF_LOW, FALLOFF_LOW_NAME, TROE_FALLOFF, TROE_F_ALPHA, TROE_F_TS, TROE_F_TSS, TROE_F_TSSS } |
| enum | ParsingUnit { MOL_WEIGHT = 0, MASS_ENTHALPY } |
Functions | |
| template<typename CoeffType , typename VectorCoeffType > | |
| KineticsType< CoeffType, VectorCoeffType > * | build_rate (const VectorCoeffType &data, const KineticsModel::KineticsModel &kin) |
| cross-road to kinetics model More... | |
| template<typename CoeffType , typename VectorCoeffType , typename VectorType > | |
| void | reset_rate (KineticsType< CoeffType, VectorCoeffType > &kin, const VectorType &coefs) |
| template<typename CoeffType , typename VectorCoeffType > | |
| void | reset_parameter_of_rate (KineticsType< CoeffType, VectorCoeffType > &rate, KineticsModel::Parameters parameter, const CoeffType new_value, const std::string &unit="SI") |
The rate constant models derived from the Arrhenius model have an activation energy which is stored and used, for efficiency reasons, in the reduced 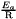 form in K. More... form in K. More... | |
| template<typename CoeffType , typename VectorCoeffType > | |
| void | reset_parameter_of_rate (KineticsType< CoeffType, VectorCoeffType > &rate, KineticsModel::Parameters parameter, const CoeffType new_value, int l, const std::string &unit="SI") |
| template<typename CoeffType > | |
| Reaction< CoeffType > * | build_reaction (const unsigned int n_species, const std::string &equation, const bool &reversible, const ReactionType::ReactionType &type, const KineticsModel::KineticsModel &kin) |
| template<class NumericType > | |
| void | read_blottner_data_ascii (MixtureViscosity< BlottnerViscosity< NumericType >, NumericType > &mu, const std::string &filename) |
| template<class NumericType > | |
| void | read_blottner_data_ascii_default (MixtureViscosity< BlottnerViscosity< NumericType >, NumericType > &mu) |
| template<class NumericType > | |
| void | read_cea_mixture_data_ascii (CEAThermoMixture< NumericType > &thermo, const std::string &filename) |
| template<class NumericType > | |
| void | read_cea_mixture_data_ascii_default (CEAThermoMixture< NumericType > &thermo) |
| template<class NumericType > | |
| void | read_cea_mixture_data_ascii (CEAThermodynamics< NumericType > &thermo, const std::string &filename) |
| template<class NumericType > | |
| void | read_cea_mixture_data (CEAThermoMixture< NumericType > &thermo, const std::string &filename, ParsingType type, bool verbose=true) |
| template<class NumericType > | |
| void | read_cea_thermo_data_ascii (CEAThermodynamics< NumericType > &thermo, const std::string &filename) |
| template<class NumericType > | |
| void | build_constant_lewis_diffusivity (MixtureDiffusion< ConstantLewisDiffusivity< NumericType >, NumericType > &D, const NumericType Le) |
| template<class NumericType > | |
| void | build_constant_lewis_diffusivity (MixtureDiffusion< ConstantLewisDiffusivity< NumericType >, NumericType > &D, const std::vector< NumericType > &Le) |
| template<class MicroThermo , class NumericType > | |
| void | build_eucken_thermal_conductivity (MixtureConductivity< EuckenThermalConductivity< MicroThermo >, NumericType > &k, const MicroThermo &thermo) |
| template<class MicroThermo , class NumericType > | |
| void | build_kinetics_theory_thermal_conductivity (MixtureConductivity< KineticsTheoryThermalConductivity< MicroThermo, NumericType >, NumericType > &k, const MicroThermo &thermo) |
| template<class NumericType > | |
| void | read_nasa_mixture_data_ascii (NASAThermoMixture< NumericType, NASA7CurveFit< NumericType > > &thermo, const std::string &filename) |
| template<class NumericType , typename CurveType > | |
| void | read_nasa_mixture_data (NASAThermoMixture< NumericType, CurveType > &thermo, const std::string &filename=DefaultSourceFilename::thermo_data(), ParsingType=ASCII, bool verbose=true) |
| template<class NumericType > | |
| void | read_reaction_set_data_xml (const std::string &filename, const bool verbose, ReactionSet< NumericType > &reaction_set) |
| template<class NumericType > | |
| void | read_reaction_set_data_chemkin (const std::string &filename, const bool verbose, ReactionSet< NumericType > &reaction_set) |
| template<typename NumericType > | |
| void | read_reaction_set_data (const std::string &filename, const bool verbose, ReactionSet< NumericType > &reaction_set, ParsingType type=ASCII) |
| template<typename NumericType > | |
| void | verify_unit_of_parameter (Units< NumericType > &default_unit, const std::string &provided_unit, const std::vector< std::string > &accepted_unit, const std::string &equation, const std::string ¶meter_name) |
| template<typename NumericType > | |
| void | read_chemical_species_composition (ParserBase< NumericType > *parser, ChemicalMixture< NumericType > &mixture) |
| template<class NumericType > | |
| void | read_species_data (ParserBase< NumericType > *parser, ChemicalMixture< NumericType > &chem_mixture) |
| template<class NumericType > | |
| void | read_species_vibrational_data (ParserBase< NumericType > *parser, ChemicalMixture< NumericType > &chem_mixture) |
| template<class NumericType > | |
| void | read_species_electronic_data (ParserBase< NumericType > *parser, ChemicalMixture< NumericType > &chem_mixture) |
| template<class NumericType > | |
| void | read_sutherland_data_ascii (MixtureViscosity< SutherlandViscosity< NumericType >, NumericType > &mu, const std::string &filename) |
| template<class NumericType > | |
| void | read_sutherland_data_ascii_default (MixtureViscosity< SutherlandViscosity< NumericType >, NumericType > &mu) |
| template<typename NumericType > | |
| void | read_transport_species_data_ascii (TransportMixture< NumericType > &transport, const std::string &filename) |
| template<typename NumericType > | |
| void | read_transport_species_data (ParserBase< NumericType > *parser, TransportMixture< NumericType > &transport) |
| ANTIOCH_NUMERIC_TYPE_CLASS_INSTANTIATE (ASCIIParser) | |
| ANTIOCH_ASCII_PARSER_INSTANTIATE () | |
| ANTIOCH_BLOTTNER_PARSING_INSTANTIATE () | |
| ANTIOCH_CEA_MIXTURE_DATA_ASCII_PARSING_INSTANTIATE () | |
| template void | read_cea_mixture_data< float > (CEAThermoMixture< float > &, const std::string &, ParsingType, bool) |
| template void | read_cea_mixture_data< double > (CEAThermoMixture< double > &, const std::string &, ParsingType, bool) |
| template void | read_cea_mixture_data< long double > (CEAThermoMixture< long double > &, const std::string &, ParsingType, bool) |
| ANTIOCH_NASA_MIXTURE_PARSING_INSTANTIATE (NASA7CurveFit) | |
| ANTIOCH_NASA_MIXTURE_PARSING_INSTANTIATE (NASA9CurveFit) | |
| ANTIOCH_NASA_MIXTURE_PARSING_INSTANTIATE (CEACurveFit) | |
| template void | read_nasa_mixture_data_ascii< float > (NASAThermoMixture< float, NASA7CurveFit< float > > &, const std::string &) |
| template void | read_nasa_mixture_data_ascii< double > (NASAThermoMixture< double, NASA7CurveFit< double > > &, const std::string &) |
| template void | read_nasa_mixture_data_ascii< long double > (NASAThermoMixture< long double, NASA7CurveFit< long double > > &, const std::string &) |
| ANTIOCH_READ_REACTION_SET_DATA_INSTANTIATE () | |
| ANTIOCH_SPECIES_PARSING_INSTANTIATE () | |
| ANTIOCH_SUTHERLAND_PARSING_INSTANTIATE () | |
| template void | read_transport_species_data< float > (ParserBase< float > *, TransportMixture< float > &) |
| template void | read_transport_species_data< double > (ParserBase< double > *, TransportMixture< double > &) |
| template void | read_transport_species_data< long double > (ParserBase< long double > *, TransportMixture< long double > &) |
| template void | read_transport_species_data_ascii< float > (TransportMixture< float > &, const std::string &) |
| template void | read_transport_species_data_ascii< double > (TransportMixture< double > &, const std::string &) |
| template void | read_transport_species_data_ascii< long double > (TransportMixture< long double > &, const std::string &) |
| ANTIOCH_NUMERIC_TYPE_CLASS_INSTANTIATE (XMLParser) | |
| ANTIOCH_XML_PARSER_INSTANTIATE () | |
| template<typename StateType > | |
| const ANTIOCH_AUTO(StateType) EuckenThermalConductivity < ThermoEvaluator > return | trans (s, mu_s) |
| template<typename StateType > | |
| const | ANTIOCH_AUTO (StateType) KineticsTheoryThermalConductivity< ThermoEvaluator |
| template<typename T > | |
| Antioch::enable_if_c< is_eigen < T >::value, typename value_type< T >::type >::type | max (const T &in) |
| template<typename T > | |
| Antioch::enable_if_c< is_eigen < T >::value, typename value_type< T >::type >::type | min (const T &in) |
| template<template< typename, int, int, int, int, int > class _Matrix, typename _Scalar , int _Rows, int _Cols, int _Options, int _MaxRows, int _MaxCols> | |
| _Matrix< _Scalar, _Rows, _Cols, _Options, _MaxRows, _MaxCols > | zero_clone (const _Matrix< _Scalar, _Rows, _Cols, _Options, _MaxRows, _MaxCols > &ex) |
| template<template< typename, int, int, int, int, int > class _Matrix, typename _Scalar , int _Rows, int _Cols, int _Options, int _MaxRows, int _MaxCols, typename Scalar2 > | |
| void | zero_clone (_Matrix< _Scalar, _Rows, _Cols, _Options, _MaxRows, _MaxCols > &output, const _Matrix< Scalar2, _Rows, _Cols, _Options, _MaxRows, _MaxCols > &ex) |
| template<template< typename, int, int, int, int, int > class _Matrix, typename _Scalar , int _Rows, int _Cols, int _Options, int _MaxRows, int _MaxCols, typename Scalar > | |
| _Matrix< _Scalar, _Rows, _Cols, _Options, _MaxRows, _MaxCols > | constant_clone (const _Matrix< _Scalar, _Rows, _Cols, _Options, _MaxRows, _MaxCols > &ex, const Scalar &value) |
| template<template< typename, int, int, int, int, int > class _Matrix, typename _Scalar , int _Rows, int _Cols, int _Options, int _MaxRows, int _MaxCols, typename Scalar > | |
| void | constant_fill (_Matrix< _Scalar, _Rows, _Cols, _Options, _MaxRows, _MaxCols > &output, const Scalar &value) |
| template<template< typename, int, int, int, int, int > class _Matrix, typename _Scalar , int _Rows, int _Cols, int _Options, int _MaxRows, int _MaxCols> | |
| void | set_zero (_Matrix< _Scalar, _Rows, _Cols, _Options, _MaxRows, _MaxCols > &a) |
| template<typename Condition , typename T1 , typename T2 > | |
| enable_if_c< is_eigen< T1 > ::value &&is_eigen< T2 > ::value, typename state_type < T1 >::type >::type | if_else (const Condition &condition, const T1 &if_true, const T2 &if_false) |
| template<typename VectorT , template< typename, int, int, int, int, int > class _Matrix, typename _UIntT , int _Rows, int _Cols, int _Options, int _MaxRows, int _MaxCols> | |
| enable_if_c< is_eigen < typename value_type< VectorT > ::type >::value, typename value_type< VectorT >::type > ::type | eval_index (const VectorT &vec, const _Matrix< _UIntT, _Rows, _Cols, _Options, _MaxRows, _MaxCols > &index) |
| template<typename T > | |
| bool | conjunction_root (const T &vec, eigen_library_tag) |
| template<typename T > | |
| bool | disjunction_root (const T &vec, eigen_library_tag) |
| void | skip_comment_lines (std::istream &in, const char comment_start) |
| Skip comment lines in the header of an ASCII text file prefixed with the comment character 'comment_start'. More... | |
| template<typename T > | |
| Antioch::enable_if_c < is_metaphysicl< T >::value, typename value_type< T >::type > ::type | max (const T &in) |
| template<typename T > | |
| Antioch::enable_if_c < is_metaphysicl< T >::value, typename value_type< T >::type > ::type | min (const T &in) |
| template<typename Tbool , typename Ttrue , typename Tfalse > | |
| Antioch::enable_if_c < is_metaphysicl< Tbool > ::value &&is_metaphysicl < Ttrue >::value &&is_metaphysicl< Tfalse > ::value, typename state_type < Ttrue >::type >::type | if_else (const Tbool &condition, const Ttrue &if_true, const Tfalse &if_false) |
| template<typename VectorT , typename UIntType > | |
| Antioch::enable_if_c < is_metaphysicl< typename Antioch::value_type< VectorT > ::type >::value &&is_metaphysicl< UIntType > ::value, typename Antioch::value_type< VectorT > ::type >::type | eval_index (const VectorT &vec, const UIntType &indexes) |
| ANTIOCH_PLAIN_SCALAR (float) | |
| ANTIOCH_PLAIN_SCALAR (double) | |
| ANTIOCH_PLAIN_SCALAR (long double) | |
| ANTIOCH_PLAIN_SCALAR (short) | |
| ANTIOCH_PLAIN_SCALAR (int) | |
| ANTIOCH_PLAIN_SCALAR (unsigned short) | |
| template<typename T > | |
| void | set_zero (T &output) |
| template<typename T > | |
| T | zero_clone (const T &) |
| template<typename T , typename T2 > | |
| void | zero_clone (T &output, const T2 &) |
| template<typename T > | |
| void | init_clone (T &output, const T &example) |
| template<typename Vector , typename Scalar > | |
| void | init_constant (Vector &output, const Scalar &example) |
| template<typename T , typename Scalar > | |
| T | constant_clone (const T &, const Scalar &value) |
| template<typename T , typename Scalar > | |
| void | constant_fill (T &output, const Scalar &value) |
| template<typename T , typename VectorScalar > | |
| T | custom_clone (const T &, const VectorScalar &values, unsigned int indexes) |
| template<typename T , typename VectorScalar > | |
| T | custom_clone (const T &, const VectorScalar &values, const typename Antioch::rebind< T, unsigned int >::type &indexes) |
| template<typename VectorT > | |
| value_type< VectorT >::type | eval_index (const VectorT &vec, unsigned int index) |
| bool | conjunction (const bool &vec) |
| template<typename T > | |
| bool | conjunction (const T &vec) |
| template<typename T > | |
| bool | conjunction_root (const T &vec, numeric_library_tag) |
| bool | disjunction (const bool &vec) |
| template<typename T > | |
| bool | disjunction (const T &vec) |
| template<typename T > | |
| bool | disjunction_root (const T &vec, numeric_library_tag) |
| template<typename T1 , typename T2 > | |
| void | zero_clone (T1 &output, const T2 &example) |
| template<typename T > | |
| T | if_else (bool condition, T if_true, T if_false) |
| void | split_string (const std::string &input, const std::string &delimiter, std::vector< std::string > &results) |
| All characters in delimiter will be treated as a delimiter. More... | |
| void | remove_newline_from_strings (std::vector< std::string > &strings) |
| Strips newline characters from strings in the input vector, strings. More... | |
| template<typename T > | |
| T | string_to_T (const std::string &input) |
| template<typename Type > | |
| std::pair< std::string, Type > | split_string_on_colon (const std::string &token) |
| int | SplitString (const std::string &input, const std::string &delimiter, std::vector< std::string > &results, bool includeEmpties=true) |
| Taken from FIN-S for XML parsing. More... | |
| int | ascii_getline (std::istream &buf, std::string &line) |
| adapted getline, never believe ascii file for the formatting of end-of-line. More... | |
| KineticsModel::Parameters | string_to_kin_enum (const std::string &str) |
| ReactionType::Parameters | string_to_chem_enum (const std::string &str) |
| template<typename T > | |
| T | max (const std::valarray< T > &in) |
| template<typename T > | |
| T | min (const std::valarray< T > &in) |
| template<typename T > | |
| std::valarray< T > | zero_clone (const std::valarray< T > &example) |
| template<typename T1 , typename T2 > | |
| void | zero_clone (std::valarray< T1 > &output, const std::valarray< T2 > &example) |
| template<typename T , typename Scalar > | |
| std::valarray< T > | constant_clone (const std::valarray< T > &example, const Scalar &value) |
| template<typename T > | |
| void | init_clone (std::valarray< T > &output, const std::valarray< T > &example) |
| template<typename T > | |
| std::valarray< T > | if_else (const std::valarray< bool > &condition, const std::valarray< T > &if_true, const std::valarray< T > &if_false) |
| template<typename VectorT > | |
| Antioch::enable_if_c < Antioch::is_valarray < typename value_type< VectorT > ::type >::value, typename value_type< VectorT >::type > ::type | eval_index (const VectorT &vec, const std::valarray< unsigned int > &index) |
| template<typename T > | |
| std::vector< T > | zero_clone (const std::vector< T > &example) |
| template<typename T1 , typename T2 > | |
| void | zero_clone (std::vector< T1 > &output, const std::vector< T2 > &example) |
| template<typename T , typename Scalar > | |
| std::vector< T > | zero_clone (const std::vector< T > &example, const Scalar &value) |
| template<typename T > | |
| void | set_zero (std::vector< T > &a) |
| template<typename T , typename VectorScalar > | |
| std::vector< T > | custom_clone (const std::vector< T > &, const VectorScalar &vecsrc, std::vector< unsigned int > &indexes) |
| template<typename T , typename Scalar > | |
| std::vector< T > | constant_clone (const std::vector< T > &example, const Scalar &value) |
| template<typename T , typename VectorScalar > | |
| std::vector< T > | custom_clone (const std::vector< T > &vec, const VectorScalar &vecsrc, const std::vector< unsigned int > &indexes) |
| template<typename T > | |
| T | max (const vex::vector< T > &in) |
| template<typename T > | |
| T | min (const vex::vector< T > &in) |
| template<typename T > | |
| vex::vector< T > | zero_clone (const vex::vector< T > &example) |
| template<typename T1 , typename T2 > | |
| void | zero_clone (vex::vector< T1 > &output, const vex::vector< T2 > &example) |
| template<typename T , typename Scalar > | |
| vex::vector< T > | constant_clone (const vex::vector< T > &example, const Scalar &value) |
| template<typename T > | |
| void | init_clone (vex::vector< T > &output, const vex::vector< T > &example) |
| template<typename BoolInput , typename IfValue , typename ElseValue > | |
| boost::proto::result_of::make_expr < boost::proto::tag::if_else_, boost::proto::deduce_domain, const vex::vector_expression < BoolInput > &, const IfValue &, const ElseValue & >::type const | if_else (const vex::vector_expression< BoolInput > &condition, const IfValue &if_true, const ElseValue &if_false) |
| template<typename T > | |
| bool | disjunction_root (const T &vec_input, vexcl_library_tag) |
| template<typename T > | |
| bool | conjunction_root (const T &vec_input, vexcl_library_tag) |
| template<typename VectorT , typename IntT > | |
| enable_if_c < vex::is_vector_expression < typename value_type< VectorT > ::type >::value &&vex::is_vector_expression < IntT >::value, typename value_type< VectorT >::type > ::type | eval_index (const VectorT &vec, const IntT &index) |
| int | get_antioch_version () |
Detailed Description
The parameters are reduced parameters.
Everything here is deprecated, it is for backward compatibility, using the deprecated name/object CEAThermo...<NumericType>.
The complete Van't Hoff equation is (all the others are deductible from it) 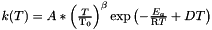 with
with 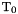 being a reference temperature and
being a reference temperature and 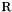 the ideal gas constant. The reduced parameters are:
the ideal gas constant. The reduced parameters are:
![\[ \begin{array}{ccc}\toprule \text{Parameter} & \text{reduced parameter} \\\midrule A & A \mathrm{T_0}^\beta \\ \beta & \beta \\ E_a & \frac{E_a}{\mathrm{R}}\\ D & D\\\bottomrule \end{array} \]](form_46.png)
This library is intended for performances, thus the reference temperature is taken equal to one in order to skip the division step ( 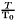 ). This is not an option, this is an obligation: the kinetics equations are coded without the division step.
). This is not an option, this is an obligation: the kinetics equations are coded without the division step.
We required to provide:
for both descriptions (Mixture, dynamics).
Typedef Documentation
Definition at line 106 of file default_filename.h.
| typedef unsigned int Antioch::Species |
Definition at line 49 of file chemical_mixture.h.
Enumeration Type Documentation
| enum Antioch::ParsingKey |
Definition at line 36 of file parsing_enum.h.
| enum Antioch::ParsingType |
| Enumerator | |
|---|---|
| ASCII | |
| XML | |
| CHEMKIN | |
Definition at line 32 of file parsing_enum.h.
| enum Antioch::ParsingUnit |
| Enumerator | |
|---|---|
| MOL_WEIGHT | |
| MASS_ENTHALPY | |
Definition at line 77 of file parsing_enum.h.
Function Documentation
| Antioch::ANTIOCH_ASCII_PARSER_INSTANTIATE | ( | ) |
|
inline |
| Antioch::ANTIOCH_BLOTTNER_PARSING_INSTANTIATE | ( | ) |
| Antioch::ANTIOCH_CEA_MIXTURE_DATA_ASCII_PARSING_INSTANTIATE | ( | ) |
| Antioch::ANTIOCH_NASA_MIXTURE_PARSING_INSTANTIATE | ( | NASA7CurveFit | ) |
| Antioch::ANTIOCH_NASA_MIXTURE_PARSING_INSTANTIATE | ( | NASA9CurveFit | ) |
| Antioch::ANTIOCH_NASA_MIXTURE_PARSING_INSTANTIATE | ( | CEACurveFit | ) |
| Antioch::ANTIOCH_NUMERIC_TYPE_CLASS_INSTANTIATE | ( | ASCIIParser | ) |
| Antioch::ANTIOCH_NUMERIC_TYPE_CLASS_INSTANTIATE | ( | XMLParser | ) |
| Antioch::ANTIOCH_PLAIN_SCALAR | ( | float | ) |
| Antioch::ANTIOCH_PLAIN_SCALAR | ( | double | ) |
| Antioch::ANTIOCH_PLAIN_SCALAR | ( | long | double | ) |
| Antioch::ANTIOCH_PLAIN_SCALAR | ( | short | ) |
| Antioch::ANTIOCH_PLAIN_SCALAR | ( | int | ) |
| Antioch::ANTIOCH_PLAIN_SCALAR | ( | unsigned | short | ) |
| Antioch::ANTIOCH_READ_REACTION_SET_DATA_INSTANTIATE | ( | ) |
| Antioch::ANTIOCH_SPECIES_PARSING_INSTANTIATE | ( | ) |
| Antioch::ANTIOCH_SUTHERLAND_PARSING_INSTANTIATE | ( | ) |
| Antioch::ANTIOCH_XML_PARSER_INSTANTIATE | ( | ) |
|
inline |
adapted getline, never believe ascii file for the formatting of end-of-line.
end-of-line triggered by
or
- Windows
- Unix/Linux
- Mac
Definition at line 169 of file string_utils.h.
Referenced by Antioch::ChemKinParser< NumericType >::initialize(), Antioch::ChemKinParser< NumericType >::next_meaningful_line(), Antioch::ChemKinParser< NumericType >::read_thermodynamic_data_root(), and Antioch::ChemKinParser< NumericType >::species_list().
| void Antioch::build_constant_lewis_diffusivity | ( | MixtureDiffusion< ConstantLewisDiffusivity< NumericType >, NumericType > & | D, |
| const NumericType | Le | ||
| ) |
Definition at line 46 of file constant_lewis_diffusivity_building.h.
References Antioch::KineticsModel::D.
| void Antioch::build_constant_lewis_diffusivity | ( | MixtureDiffusion< ConstantLewisDiffusivity< NumericType >, NumericType > & | D, |
| const std::vector< NumericType > & | Le | ||
| ) |
Definition at line 56 of file constant_lewis_diffusivity_building.h.
References Antioch::KineticsModel::D.
| void Antioch::build_eucken_thermal_conductivity | ( | MixtureConductivity< EuckenThermalConductivity< MicroThermo >, NumericType > & | k, |
| const MicroThermo & | thermo | ||
| ) |
Definition at line 42 of file eucken_thermal_conductivity_building.h.
| void Antioch::build_kinetics_theory_thermal_conductivity | ( | MixtureConductivity< KineticsTheoryThermalConductivity< MicroThermo, NumericType >, NumericType > & | k, |
| const MicroThermo & | thermo | ||
| ) |
Definition at line 48 of file kinetics_theory_thermal_conductivity_building.h.
|
inline |
cross-road to kinetics model
We take here the parameters as:
Definition at line 89 of file kinetics_parsing.h.
References antioch_assert_equal_to, antioch_error, Antioch::KineticsModel::ARRHENIUS, Antioch::KineticsModel::BERTHELOT, Antioch::KineticsModel::BHE, Antioch::KineticsModel::CONSTANT, Antioch::KineticsModel::HERCOURT_ESSEN, Antioch::KineticsModel::KOOIJ, Antioch::KineticsModel::PHOTOCHEM, std::pow(), and Antioch::KineticsModel::VANTHOFF.
|
inline |
Definition at line 53 of file reaction_parsing.h.
References antioch_error, Antioch::ReactionType::DUPLICATE, Antioch::ReactionType::ELEMENTARY, Antioch::ReactionType::LINDEMANN_FALLOFF, Antioch::ReactionType::LINDEMANN_FALLOFF_THREE_BODY, Antioch::ReactionType::THREE_BODY, Antioch::ReactionType::TROE_FALLOFF, and Antioch::ReactionType::TROE_FALLOFF_THREE_BODY.
|
inline |
Definition at line 161 of file metaprogramming.h.
Referenced by Antioch::SigmaBinConverter< VectorCoeffType >::y_on_custom_grid().
|
inline |
Definition at line 168 of file metaprogramming.h.
References conjunction_root().
|
inline |
Definition at line 176 of file metaprogramming.h.
|
inline |
|
inline |
Definition at line 278 of file eigen_utils.h.
Referenced by conjunction().
|
inline |
|
inline |
Definition at line 128 of file metaprogramming.h.
|
inline |
Definition at line 176 of file vexcl_utils.h.
|
inline |
Definition at line 181 of file eigen_utils.h.
Referenced by Antioch::MixtureAveragedTransportEvaluator< Diffusion, Viscosity, ThermalConductivity, CoeffType >::compute_mu_mu_sqrt(), Antioch::Reaction< CoeffType, VectorCoeffType >::compute_rate_of_progress_and_derivatives(), Antioch::Converter< T >::Converter(), Antioch::NASAEvaluator< CoeffType, NASAFit >::cp(), Antioch::CEAThermodynamics< CoeffType >::cp(), Antioch::SigmaBinConverter< VectorCoeffType >::custom_bin_value(), Antioch::MixtureAveragedTransportEvaluator< Diffusion, Viscosity, ThermalConductivity, CoeffType >::diffusion_mixing_rule(), Antioch::LindemannFalloff< CoeffType >::F_and_derivatives(), Antioch::TroeFalloff< CoeffType >::F_and_derivatives(), Antioch::NASACurveFitBase< CoeffType >::interval(), Antioch::LindemannFalloff< CoeffType >::operator()(), Antioch::TroeFalloff< CoeffType >::operator()(), Antioch::Converter< T >::operator=(), Antioch::LindemannFalloff< CoeffType >::value(), and Antioch::SigmaBinConverter< VectorCoeffType >::y_on_custom_grid().
|
inline |
Definition at line 187 of file valarray_utils.h.
|
inline |
Definition at line 132 of file metaprogramming.h.
|
inline |
Definition at line 204 of file eigen_utils.h.
Referenced by Antioch::ConstantRate< CoeffType >::rate_and_derivative().
|
inline |
|
inline |
Definition at line 136 of file metaprogramming.h.
Referenced by Antioch::SigmaBinConverter< VectorCoeffType >::custom_bin_value().
|
inline |
Definition at line 140 of file metaprogramming.h.
|
inline |
Definition at line 175 of file vector_utils.h.
|
inline |
Definition at line 186 of file metaprogramming.h.
Referenced by Antioch::SigmaBinConverter< VectorCoeffType >::custom_bin_value().
|
inline |
Definition at line 193 of file metaprogramming.h.
References disjunction_root().
|
inline |
Definition at line 201 of file metaprogramming.h.
|
inline |
|
inline |
Definition at line 285 of file eigen_utils.h.
Referenced by disjunction().
|
inline |
Definition at line 154 of file metaphysicl_utils.h.
|
inline |
Definition at line 155 of file metaprogramming.h.
|
inline |
Definition at line 248 of file vexcl_utils.h.
References zero_clone().
|
inline |
Definition at line 249 of file valarray_utils.h.
|
inline |
Definition at line 268 of file eigen_utils.h.
Referenced by Antioch::SigmaBinConverter< VectorCoeffType >::y_on_custom_grid().
| int Antioch::get_antioch_version | ( | ) |
Definition at line 31 of file antioch_version.C.
|
inline |
Definition at line 133 of file metaphysicl_utils.h.
References antioch_assert_equal_to.
|
inline |
Definition at line 202 of file vexcl_utils.h.
|
inline |
|
inline |
Definition at line 226 of file valarray_utils.h.
References antioch_assert_equal_to.
|
inline |
Definition at line 250 of file eigen_utils.h.
Referenced by Antioch::NASAEvaluator< CoeffType, NASAFit >::cp(), Antioch::CEAThermodynamics< CoeffType >::cp(), Antioch::NASA7CurveFit< CoeffType >::cp_over_R(), Antioch::NASA9CurveFit< CoeffType >::cp_over_R(), Antioch::CEAThermodynamics< CoeffType >::cp_over_R(), Antioch::SigmaBinConverter< VectorCoeffType >::custom_bin_value(), Antioch::NASA7CurveFit< CoeffType >::dh_RT_minus_s_R_dT(), Antioch::NASA9CurveFit< CoeffType >::dh_RT_minus_s_R_dT(), Antioch::CEAThermodynamics< CoeffType >::dh_RT_minus_s_R_dT(), Antioch::NASA7CurveFit< CoeffType >::h_over_RT(), Antioch::NASA9CurveFit< CoeffType >::h_over_RT(), Antioch::NASA7CurveFit< CoeffType >::h_RT_minus_s_R(), Antioch::NASA9CurveFit< CoeffType >::h_RT_minus_s_R(), Antioch::CEAThermodynamics< CoeffType >::h_RT_minus_s_R(), Antioch::NASACurveFitBase< CoeffType >::interval(), Antioch::NASA7CurveFit< CoeffType >::s_over_R(), Antioch::NASA9CurveFit< CoeffType >::s_over_R(), vectester(), and Antioch::SigmaBinConverter< VectorCoeffType >::y_on_custom_grid().
|
inline |
Definition at line 115 of file metaprogramming.h.
Referenced by init_clone(), init_constant(), Antioch::SIPrefixes< T >::operator=(), and Antioch::Units< T >::set_SI_converter().
|
inline |
Definition at line 186 of file vexcl_utils.h.
|
inline |
Definition at line 200 of file valarray_utils.h.
References init_clone().
|
inline |
Definition at line 119 of file metaprogramming.h.
References init_clone().
Referenced by Antioch::WilkeEvaluator< MixtureViscosity, ThermalConductivity, CoeffType >::compute_phi(), Antioch::MixtureAveragedTransportEvaluator< Diffusion, Viscosity, ThermalConductivity, CoeffType >::D(), Antioch::MixtureAveragedTransportEvaluator< Diffusion, Viscosity, ThermalConductivity, CoeffType >::k(), Antioch::MixtureAveragedTransportEvaluator< Diffusion, Viscosity, ThermalConductivity, CoeffType >::mu(), Antioch::MixtureAveragedTransportEvaluator< Diffusion, Viscosity, ThermalConductivity, CoeffType >::mu_and_k(), Antioch::MixtureAveragedTransportEvaluator< Diffusion, Viscosity, ThermalConductivity, CoeffType >::mu_and_k_and_D(), tester(), vectester(), and Antioch::SigmaBinConverter< VectorCoeffType >::y_on_custom_grid().
|
inline |
Definition at line 67 of file metaphysicl_utils.h.
References std::max(), and max().
|
inline |
Definition at line 88 of file eigen_utils.h.
Referenced by Antioch::NASA7CurveFit< CoeffType >::cp_over_R(), Antioch::NASA9CurveFit< CoeffType >::cp_over_R(), Antioch::CEAThermodynamics< CoeffType >::cp_over_R(), Antioch::SigmaBinConverter< VectorCoeffType >::custom_bin_value(), Antioch::StatMechThermodynamics< CoeffType >::cv_rot(), Antioch::StatMechThermodynamics< CoeffType >::cv_rot_over_R(), Antioch::NASA7CurveFit< CoeffType >::dh_RT_minus_s_R_dT(), Antioch::NASA9CurveFit< CoeffType >::dh_RT_minus_s_R_dT(), Antioch::CEAThermodynamics< CoeffType >::dh_RT_minus_s_R_dT(), Antioch::StockmayerPotential< CoeffType >::extrapolate_to(), Antioch::NASA7CurveFit< CoeffType >::h_over_RT(), Antioch::NASA9CurveFit< CoeffType >::h_over_RT(), Antioch::NASA7CurveFit< CoeffType >::h_RT_minus_s_R(), Antioch::NASA9CurveFit< CoeffType >::h_RT_minus_s_R(), Antioch::CEAThermodynamics< CoeffType >::h_RT_minus_s_R(), max(), Antioch::NASA7CurveFit< CoeffType >::s_over_R(), Antioch::NASA9CurveFit< CoeffType >::s_over_R(), Antioch::StatMechThermodynamics< CoeffType >::T_from_e_tot(), test_cp(), test_val(), test_viscosity(), vec_compare(), and vectester().
|
inline |
Definition at line 109 of file vexcl_utils.h.
|
inline |
Definition at line 119 of file valarray_utils.h.
|
inline |
Definition at line 84 of file metaphysicl_utils.h.
References std::min(), and min().
|
inline |
Definition at line 98 of file eigen_utils.h.
Referenced by Antioch::NASA7CurveFit< CoeffType >::cp_over_R(), Antioch::NASA9CurveFit< CoeffType >::cp_over_R(), Antioch::CEAThermodynamics< CoeffType >::cp_over_R(), Antioch::SigmaBinConverter< VectorCoeffType >::custom_bin_value(), Antioch::NASA7CurveFit< CoeffType >::dh_RT_minus_s_R_dT(), Antioch::NASA9CurveFit< CoeffType >::dh_RT_minus_s_R_dT(), Antioch::CEAThermodynamics< CoeffType >::dh_RT_minus_s_R_dT(), Antioch::NASA7CurveFit< CoeffType >::h_over_RT(), Antioch::NASA9CurveFit< CoeffType >::h_over_RT(), Antioch::NASA7CurveFit< CoeffType >::h_RT_minus_s_R(), Antioch::NASA9CurveFit< CoeffType >::h_RT_minus_s_R(), Antioch::CEAThermodynamics< CoeffType >::h_RT_minus_s_R(), min(), Antioch::NASA7CurveFit< CoeffType >::s_over_R(), Antioch::NASA9CurveFit< CoeffType >::s_over_R(), and Antioch::StatMechThermodynamics< CoeffType >::T_from_e_tot().
|
inline |
Definition at line 118 of file vexcl_utils.h.
|
inline |
Definition at line 127 of file valarray_utils.h.
| void Antioch::read_blottner_data_ascii | ( | MixtureViscosity< BlottnerViscosity< NumericType >, NumericType > & | mu, |
| const std::string & | filename | ||
| ) |
Definition at line 41 of file blottner_parsing.C.
References antioch_error, Antioch::ChemicalMixture< CoeffType >::chemical_mixture(), skip_comment_lines(), and Antioch::ChemicalMixture< CoeffType >::species_name_map().
Referenced by read_blottner_data_ascii_default(), and tester().
| void Antioch::read_blottner_data_ascii_default | ( | MixtureViscosity< BlottnerViscosity< NumericType >, NumericType > & | mu | ) |
Definition at line 90 of file blottner_parsing.C.
References Antioch::DefaultSourceFilename::blottner_data(), and read_blottner_data_ascii().
| void Antioch::read_cea_mixture_data | ( | CEAThermoMixture< NumericType > & | thermo, |
| const std::string & | filename, | ||
| ParsingType | type, | ||
| bool | verbose = true |
||
| ) |
Definition at line 41 of file cea_mixture_parsing.C.
References antioch_error, antioch_parsing_error, ASCII, Antioch::NASAThermoMixture< CoeffType, CEACurveFit< CoeffType > >::check(), CHEMKIN, Antioch::ParserBase< NumericType >::read_thermodynamic_data(), and XML.
Referenced by read_cea_mixture_data_ascii(), read_cea_mixture_data_ascii_default(), and tester().
| template void Antioch::read_cea_mixture_data< double > | ( | CEAThermoMixture< double > & | , |
| const std::string & | , | ||
| ParsingType | , | ||
| bool | |||
| ) |
| template void Antioch::read_cea_mixture_data< float > | ( | CEAThermoMixture< float > & | , |
| const std::string & | , | ||
| ParsingType | , | ||
| bool | |||
| ) |
| template void Antioch::read_cea_mixture_data< long double > | ( | CEAThermoMixture< long double > & | , |
| const std::string & | , | ||
| ParsingType | , | ||
| bool | |||
| ) |
| void Antioch::read_cea_mixture_data_ascii | ( | CEAThermoMixture< NumericType > & | thermo, |
| const std::string & | filename | ||
| ) |
Definition at line 45 of file cea_mixture_ascii_parsing.C.
References antioch_deprecated, ASCII, and read_cea_mixture_data().
Referenced by Antioch::CEAThermodynamics< CoeffType >::CEAThermodynamics(), and vectester().
| void Antioch::read_cea_mixture_data_ascii | ( | CEAThermodynamics< NumericType > & | thermo, |
| const std::string & | filename | ||
| ) |
Definition at line 52 of file cea_mixture_ascii_parsing.C.
References antioch_deprecated, and Antioch::ASCIIParser< NumericType >::read_thermodynamic_data().
| void Antioch::read_cea_mixture_data_ascii_default | ( | CEAThermoMixture< NumericType > & | thermo | ) |
Definition at line 38 of file cea_mixture_ascii_parsing.C.
References antioch_deprecated, ASCII, read_cea_mixture_data(), and Antioch::DefaultSourceFilename::thermo_data().
|
inline |
Definition at line 40 of file cea_thermo_ascii_parsing.C.
References Antioch::CEAThermodynamics< CoeffType >::add_curve_fit(), antioch_error, Antioch::CEAThermodynamics< CoeffType >::check(), Antioch::CEAThermodynamics< CoeffType >::chemical_mixture(), skip_comment_lines(), and Antioch::ChemicalMixture< CoeffType >::species_name_map().
| void Antioch::read_chemical_species_composition | ( | ParserBase< NumericType > * | parser, |
| ChemicalMixture< NumericType > & | mixture | ||
| ) |
Definition at line 40 of file species_parsing.C.
References Antioch::ChemicalMixture< CoeffType >::initialize_species(), and Antioch::ParserBase< NumericType >::species_list().
| void Antioch::read_nasa_mixture_data | ( | NASAThermoMixture< NumericType, CurveType > & | thermo, |
| const std::string & | filename = DefaultSourceFilename::thermo_data(), |
||
| ParsingType | type = ASCII, |
||
| bool | verbose = true |
||
| ) |
Definition at line 42 of file nasa_mixture_parsing.C.
References antioch_error, antioch_parsing_error, ASCII, Antioch::NASAThermoMixture< CoeffType, NASAFit >::check(), CHEMKIN, Antioch::ParserBase< NumericType >::read_thermodynamic_data(), and XML.
Referenced by read_nasa_mixture_data_ascii(), AntiochTesting::NASA9XMLParsingTest< long double >::test_supplied_species(), AntiochTesting::NASA7XMLParsingTest< long double >::test_supplied_species(), test_type(), and tester().
| void Antioch::read_nasa_mixture_data_ascii | ( | NASAThermoMixture< NumericType, NASA7CurveFit< NumericType > > & | thermo, |
| const std::string & | filename | ||
| ) |
Definition at line 74 of file nasa_mixture_parsing.C.
References antioch_deprecated, CHEMKIN, and read_nasa_mixture_data().
| template void Antioch::read_nasa_mixture_data_ascii< double > | ( | NASAThermoMixture< double, NASA7CurveFit< double > > & | , |
| const std::string & | |||
| ) |
| template void Antioch::read_nasa_mixture_data_ascii< float > | ( | NASAThermoMixture< float, NASA7CurveFit< float > > & | , |
| const std::string & | |||
| ) |
| template void Antioch::read_nasa_mixture_data_ascii< long double > | ( | NASAThermoMixture< long double, NASA7CurveFit< long double > > & | , |
| const std::string & | |||
| ) |
| void Antioch::read_reaction_set_data | ( | const std::string & | filename, |
| const bool | verbose, | ||
| ReactionSet< NumericType > & | reaction_set, | ||
| ParsingType | type = ASCII |
||
| ) |
Definition at line 77 of file read_reaction_set_data.C.
References Antioch::Reaction< CoeffType, VectorCoeffType >::add_forward_rate(), Antioch::Reaction< CoeffType, VectorCoeffType >::add_product(), Antioch::Reaction< CoeffType, VectorCoeffType >::add_reactant(), Antioch::ReactionSet< CoeffType >::add_reaction(), antioch_assert, antioch_assert_equal_to, antioch_do_once, antioch_error, antioch_not_implemented, antioch_parameter_required, antioch_parsing_error, Antioch::KineticsModel::ARRHENIUS, ASCII, Antioch::KineticsModel::BERTHELOT, Antioch::KineticsModel::BHE, Antioch::ReactionSet< CoeffType >::chemical_mixture(), CHEMKIN, Antioch::Units< T >::clear(), Antioch::KineticsModel::CONSTANT, Antioch::ReactionType::DUPLICATE, Antioch::ParserBase< NumericType >::efficiencies(), Antioch::ReactionType::ELEMENTARY, Antioch::Reaction< CoeffType >::equation(), Antioch::Reaction< CoeffType, VectorCoeffType >::equation(), Antioch::Units< T >::factor_to_some_unit(), Antioch::Units< T >::get_SI_factor(), Antioch::Units< T >::get_symbol(), Antioch::KineticsModel::HERCOURT_ESSEN, Antioch::ParserBase< NumericType >::initialize(), Antioch::Units< T >::is_homogeneous(), Antioch::ParserBase< NumericType >::is_k0(), Antioch::Units< T >::is_united(), Antioch::KineticsModel::KOOIJ, Antioch::ReactionType::LINDEMANN_FALLOFF, Antioch::ReactionType::LINDEMANN_FALLOFF_THREE_BODY, Antioch::Reaction< CoeffType, VectorCoeffType >::n_rate_constants(), Antioch::ChemicalMixture< CoeffType >::n_species(), Antioch::KineticsModel::PHOTOCHEM, Antioch::ParserBase< NumericType >::products_orders(), Antioch::ParserBase< NumericType >::products_pairs(), Antioch::ParserBase< NumericType >::rate_constant(), Antioch::ParserBase< NumericType >::rate_constant_activation_energy_parameter(), Antioch::ParserBase< NumericType >::rate_constant_Berthelot_coefficient_parameter(), Antioch::ParserBase< NumericType >::rate_constant_cross_section_parameter(), Antioch::ParserBase< NumericType >::rate_constant_lambda_parameter(), Antioch::ParserBase< NumericType >::rate_constant_power_parameter(), Antioch::ParserBase< NumericType >::rate_constant_preexponential_parameter(), Antioch::ParserBase< NumericType >::rate_constant_Tref_parameter(), Antioch::ParserBase< NumericType >::reactants_orders(), Antioch::ParserBase< NumericType >::reactants_pairs(), Antioch::ParserBase< NumericType >::reaction(), Antioch::ParserBase< NumericType >::reaction_chemical_process(), Antioch::ParserBase< NumericType >::reaction_equation(), Antioch::ParserBase< NumericType >::reaction_id(), Antioch::ParserBase< NumericType >::reaction_kinetics_model(), Antioch::ParserBase< NumericType >::reaction_reversible(), Antioch::Reaction< CoeffType, VectorCoeffType >::set_efficiency(), Antioch::Reaction< CoeffType, VectorCoeffType >::set_id(), Antioch::Units< T >::set_unit(), Antioch::ChemicalMixture< CoeffType >::species_name_map(), Antioch::Units< T >::substract(), Antioch::Reaction< CoeffType, VectorCoeffType >::swap_forward_rates(), Antioch::ReactionType::THREE_BODY, Antioch::ParserBase< NumericType >::Troe(), Antioch::ParserBase< NumericType >::Troe_alpha_parameter(), Antioch::ReactionType::TROE_FALLOFF, Antioch::ReactionType::TROE_FALLOFF_THREE_BODY, Antioch::ParserBase< NumericType >::Troe_T1_parameter(), Antioch::ParserBase< NumericType >::Troe_T2_parameter(), Antioch::ParserBase< NumericType >::Troe_T3_parameter(), Antioch::Reaction< CoeffType, VectorCoeffType >::type(), Antioch::KineticsModel::VANTHOFF, Antioch::ParserBase< NumericType >::verify_Kooij_in_place_of_Arrhenius(), verify_unit_of_parameter(), Antioch::ParserBase< NumericType >::where_is_k0(), and XML.
|
inline |
Definition at line 149 of file read_reaction_set_data.h.
References CHEMKIN.
|
inline |
Definition at line 140 of file read_reaction_set_data.h.
References XML.
| void Antioch::read_species_data | ( | ParserBase< NumericType > * | parser, |
| ChemicalMixture< NumericType > & | chem_mixture | ||
| ) |
Definition at line 49 of file species_parsing.C.
References antioch_error, Antioch::ChemicalMixture< CoeffType >::chemical_species(), Antioch::ParserBase< NumericType >::file(), Antioch::ParserBase< NumericType >::read_chemical_species(), Antioch::ChemicalMixture< CoeffType >::species_inverse_name_map(), and Antioch::ChemicalMixture< CoeffType >::species_list().
| void Antioch::read_species_electronic_data | ( | ParserBase< NumericType > * | parser, |
| ChemicalMixture< NumericType > & | chem_mixture | ||
| ) |
Definition at line 103 of file species_parsing.C.
References Antioch::ChemicalMixture< CoeffType >::chemical_species(), Antioch::ParserBase< NumericType >::file(), and Antioch::ParserBase< NumericType >::read_electronic_data().
| void Antioch::read_species_vibrational_data | ( | ParserBase< NumericType > * | parser, |
| ChemicalMixture< NumericType > & | chem_mixture | ||
| ) |
Definition at line 81 of file species_parsing.C.
References Antioch::ChemicalMixture< CoeffType >::chemical_species(), Antioch::ParserBase< NumericType >::file(), and Antioch::ParserBase< NumericType >::read_vibrational_data().
| void Antioch::read_sutherland_data_ascii | ( | MixtureViscosity< SutherlandViscosity< NumericType >, NumericType > & | mu, |
| const std::string & | filename | ||
| ) |
Definition at line 42 of file sutherland_parsing.C.
References antioch_error, Antioch::ChemicalMixture< CoeffType >::chemical_mixture(), skip_comment_lines(), and Antioch::ChemicalMixture< CoeffType >::species_name_map().
Referenced by read_sutherland_data_ascii_default().
| void Antioch::read_sutherland_data_ascii_default | ( | MixtureViscosity< SutherlandViscosity< NumericType >, NumericType > & | mu | ) |
Definition at line 87 of file sutherland_parsing.C.
References read_sutherland_data_ascii(), and Antioch::DefaultSourceFilename::sutherland_data().
| void Antioch::read_transport_species_data | ( | ParserBase< NumericType > * | parser, |
| TransportMixture< NumericType > & | transport | ||
| ) |
Definition at line 37 of file transport_species_parsing.C.
References Antioch::ParserBase< NumericType >::file(), Antioch::TransportMixture< CoeffType >::n_species(), Antioch::ParserBase< NumericType >::read_transport_data(), Antioch::TransportMixture< CoeffType >::species_inverse_name_map(), and Antioch::TransportMixture< CoeffType >::transport_species().
Referenced by read_transport_species_data_ascii(), and Antioch::TransportMixture< CoeffType >::TransportMixture().
| template void Antioch::read_transport_species_data< double > | ( | ParserBase< double > * | , |
| TransportMixture< double > & | |||
| ) |
| template void Antioch::read_transport_species_data< float > | ( | ParserBase< float > * | , |
| TransportMixture< float > & | |||
| ) |
| template void Antioch::read_transport_species_data< long double > | ( | ParserBase< long double > * | , |
| TransportMixture< long double > & | |||
| ) |
| void Antioch::read_transport_species_data_ascii | ( | TransportMixture< NumericType > & | transport, |
| const std::string & | filename | ||
| ) |
Definition at line 68 of file transport_species_parsing.C.
References antioch_deprecated, and read_transport_species_data().
| template void Antioch::read_transport_species_data_ascii< double > | ( | TransportMixture< double > & | , |
| const std::string & | |||
| ) |
| template void Antioch::read_transport_species_data_ascii< float > | ( | TransportMixture< float > & | , |
| const std::string & | |||
| ) |
| template void Antioch::read_transport_species_data_ascii< long double > | ( | TransportMixture< long double > & | , |
| const std::string & | |||
| ) |
| void Antioch::remove_newline_from_strings | ( | std::vector< std::string > & | strings | ) |
Strips newline characters from strings in the input vector, strings.
Note that if an element of the vector is only the newline character, that element will be removed from the vector.
Definition at line 56 of file string_utils.C.
Referenced by Antioch::XMLParser< NumericType >::read_thermodynamic_data_root(), Antioch::XMLParser< NumericType >::species_list(), and AntiochTesting::StringUtilitiesTest::test_strip_newlines().
| void Antioch::reset_parameter_of_rate | ( | KineticsType< CoeffType, VectorCoeffType > & | rate, |
| KineticsModel::Parameters | parameter, | ||
| const CoeffType | new_value, | ||
| const std::string & | unit = "SI" |
||
| ) |
The rate constant models derived from the Arrhenius model have an activation energy which is stored and used, for efficiency reasons, in the reduced 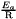 form in K.
form in K.
This calls for a subtle management of the get/set function. This is done by a unit management, by default an activation energy is an energy by quantity, so the default is to consider the SI unit for an energy, (J/mol). Therefore if the provided value is in Kelvin, it should be explicitely provided.
Definition at line 235 of file kinetics_parsing.h.
References antioch_error, Antioch::KineticsModel::ARRHENIUS, Antioch::KineticsModel::BERTHELOT, Antioch::KineticsModel::BHE, Antioch::KineticsModel::CONSTANT, Antioch::KineticsModel::E, Antioch::KineticsModel::HERCOURT_ESSEN, Antioch::KineticsModel::KOOIJ, Antioch::KineticsType< CoeffType, VectorCoeffType >::type(), and Antioch::KineticsModel::VANTHOFF.
Referenced by Antioch::Reaction< CoeffType, VectorCoeffType >::set_parameter_of_rate(), and tester().
| void Antioch::reset_parameter_of_rate | ( | KineticsType< CoeffType, VectorCoeffType > & | rate, |
| KineticsModel::Parameters | parameter, | ||
| const CoeffType | new_value, | ||
| int | l, | ||
| const std::string & | unit = "SI" |
||
| ) |
Definition at line 312 of file kinetics_parsing.h.
References antioch_error, Antioch::KineticsModel::PHOTOCHEM, and Antioch::KineticsType< CoeffType, VectorCoeffType >::type().
| void Antioch::reset_rate | ( | KineticsType< CoeffType, VectorCoeffType > & | kin, |
| const VectorType & | coefs | ||
| ) |
Definition at line 172 of file kinetics_parsing.h.
References antioch_error, Antioch::KineticsModel::ARRHENIUS, Antioch::KineticsModel::BERTHELOT, Antioch::KineticsModel::BHE, Antioch::KineticsModel::CONSTANT, Antioch::KineticsModel::HERCOURT_ESSEN, Antioch::KineticsModel::KOOIJ, Antioch::KineticsModel::PHOTOCHEM, Antioch::KineticsType< CoeffType, VectorCoeffType >::type(), and Antioch::KineticsModel::VANTHOFF.
Referenced by tester().
|
inline |
Definition at line 103 of file metaprogramming.h.
|
inline |
Definition at line 167 of file vector_utils.h.
References zero_clone().
|
inline |
Definition at line 217 of file eigen_utils.h.
References zero_clone().
Referenced by Antioch::Converter< T >::clear(), Antioch::DuplicateReaction< CoeffType >::compute_forward_rate_coefficient_and_derivatives(), Antioch::ElementaryReaction< CoeffType >::compute_forward_rate_coefficient_and_derivatives(), Antioch::KineticsEvaluator< CoeffType, StateType >::compute_mole_sources(), Antioch::KineticsEvaluator< CoeffType, StateType >::compute_mole_sources_and_derivs(), Antioch::Reaction< CoeffType, VectorCoeffType >::compute_rate_of_progress_and_derivatives(), Antioch::LindemannFalloff< CoeffType >::F_and_derivatives(), Antioch::ReactionSet< CoeffType >::get_parameter_of_reaction(), Antioch::ConstantRate< CoeffType >::rate_and_derivative(), and tester().
|
inline |
Skip comment lines in the header of an ASCII text file prefixed with the comment character 'comment_start'.
This is put in Antioch namespace so we can reuse in a few classes where we are reading in text tables. Originally taken from FIN-S.
Definition at line 46 of file input_utils.h.
Referenced by read_blottner_data_ascii(), read_cea_thermo_data_ascii(), read_sutherland_data_ascii(), and Antioch::ParserBase< NumericType >::skip_comments().
| void Antioch::split_string | ( | const std::string & | input, |
| const std::string & | delimiter, | ||
| std::vector< std::string > & | results | ||
| ) |
All characters in delimiter will be treated as a delimiter.
Definition at line 34 of file string_utils.C.
Referenced by Antioch::XMLParser< NumericType >::efficiencies(), Antioch::XMLParser< NumericType >::get_parameter(), Antioch::XMLParser< NumericType >::molecules_pairs(), Antioch::XMLParser< NumericType >::read_thermodynamic_data_root(), Antioch::XMLParser< NumericType >::species_list(), and AntiochTesting::StringUtilitiesTest::test_split_string().
|
inline |
|
inline |
Taken from FIN-S for XML parsing.
Definition at line 83 of file string_utils.h.
References std::int.
Referenced by Antioch::ChemKinParser< NumericType >::initialize(), Antioch::ChemKinParser< NumericType >::next_reaction(), Antioch::ChemKinParser< NumericType >::parse_coefficients_line(), Antioch::ChemKinParser< NumericType >::parse_equation(), Antioch::ChemKinParser< NumericType >::parse_equation_coef(), Antioch::ChemKinParser< NumericType >::parse_orders(), Antioch::ChemKinParser< NumericType >::parse_reversible_parameters(), Antioch::ChemKinParser< NumericType >::read_thermodynamic_data_root(), and Antioch::ChemKinParser< NumericType >::species_list().
|
inline |
Definition at line 232 of file string_utils.h.
References Antioch::ReactionType::EFFICIENCIES, Antioch::KineticsModel::NOT_FOUND, Antioch::ReactionType::TROE_ALPHA, Antioch::ReactionType::TROE_T1, Antioch::ReactionType::TROE_T2, and Antioch::ReactionType::TROE_T3.
Referenced by Antioch::ReactionSet< CoeffType >::get_parameter_of_reaction(), and Antioch::ReactionSet< CoeffType >::set_parameter_of_reaction().
|
inline |
Definition at line 192 of file string_utils.h.
References Antioch::KineticsModel::A, Antioch::KineticsModel::B, Antioch::KineticsModel::D, Antioch::KineticsModel::E, Antioch::KineticsModel::HIGH_PRESSURE, Antioch::KineticsModel::LAMBDA, Antioch::KineticsModel::LOW_PRESSURE, Antioch::KineticsModel::NOT_FOUND, Antioch::KineticsModel::R_SCALE, Antioch::KineticsModel::SIGMA, and Antioch::KineticsModel::T_REF.
Referenced by Antioch::ReactionSet< CoeffType >::find_kinetics_model_parameter(), Antioch::ReactionSet< CoeffType >::get_parameter_of_reaction(), and Antioch::ReactionSet< CoeffType >::set_parameter_of_reaction().
|
inline |
| const ANTIOCH_AUTO (StateType) EuckenThermalConductivity<ThermoEvaluator> return Antioch::trans | ( | s | , |
| mu_s | |||
| ) |
| void Antioch::verify_unit_of_parameter | ( | Units< NumericType > & | default_unit, |
| const std::string & | provided_unit, | ||
| const std::vector< std::string > & | accepted_unit, | ||
| const std::string & | equation, | ||
| const std::string & | parameter_name | ||
| ) |
Definition at line 44 of file read_reaction_set_data.C.
References antioch_unit_error, antioch_unit_required, Antioch::Units< T >::get_symbol(), Antioch::Units< T >::is_homogeneous(), Antioch::Units< T >::is_united(), and Antioch::Units< T >::set_unit().
Referenced by read_reaction_set_data().
|
inline |
Definition at line 107 of file metaprogramming.h.
|
inline |
Definition at line 111 of file metaprogramming.h.
|
inline |
Definition at line 134 of file vector_utils.h.
References zero_clone().
|
inline |
Definition at line 145 of file vector_utils.h.
References zero_clone().
|
inline |
Definition at line 145 of file eigen_utils.h.
References zero_clone().
Referenced by Antioch::FalloffReaction< CoeffType, FalloffType >::compute_forward_rate_coefficient(), Antioch::FalloffThreeBodyReaction< CoeffType, FalloffType >::compute_forward_rate_coefficient(), Antioch::Reaction< CoeffType, VectorCoeffType >::compute_forward_rate_coefficient(), Antioch::DuplicateReaction< CoeffType >::compute_forward_rate_coefficient_and_derivatives(), Antioch::FalloffReaction< CoeffType, FalloffType >::compute_forward_rate_coefficient_and_derivatives(), Antioch::FalloffThreeBodyReaction< CoeffType, FalloffType >::compute_forward_rate_coefficient_and_derivatives(), Antioch::Reaction< CoeffType, VectorCoeffType >::compute_rate_of_progress(), Antioch::Reaction< CoeffType, VectorCoeffType >::compute_rate_of_progress_and_derivatives(), Antioch::NASA7CurveFit< CoeffType >::cp_over_R(), Antioch::NASA9CurveFit< CoeffType >::cp_over_R(), Antioch::CEAThermodynamics< CoeffType >::cp_over_R(), Antioch::SigmaBinConverter< VectorCoeffType >::custom_bin_value(), Antioch::StatMechThermodynamics< CoeffType >::cv_el(), Antioch::StatMechThermodynamics< CoeffType >::cv_vib_over_R(), Antioch::KineticsType< CoeffType, VectorCoeffType >::derivative(), Antioch::NASA7CurveFit< CoeffType >::dh_RT_minus_s_R_dT(), Antioch::NASA9CurveFit< CoeffType >::dh_RT_minus_s_R_dT(), Antioch::CEAThermodynamics< CoeffType >::dh_RT_minus_s_R_dT(), Antioch::MixtureAveragedTransportEvaluator< Diffusion, Viscosity, ThermalConductivity, CoeffType >::diffusion_mixing_rule(), Antioch::AntiochPrivate::GSLSplinerPolicy< true >::dinterpolation(), Antioch::StatMechThermodynamics< CoeffType >::e_el(), Antioch::TroeFalloff< CoeffType >::F_and_derivatives(), Antioch::NASA7CurveFit< CoeffType >::h_over_RT(), Antioch::NASA9CurveFit< CoeffType >::h_over_RT(), Antioch::NASA7CurveFit< CoeffType >::h_RT_minus_s_R(), Antioch::NASA9CurveFit< CoeffType >::h_RT_minus_s_R(), Antioch::CEAThermodynamics< CoeffType >::h_RT_minus_s_R(), Antioch::AntiochPrivate::GSLSplinerPolicy< true >::interpolation(), Antioch::NASACurveFitBase< CoeffType >::interval(), Antioch::MixtureAveragedTransportEvaluator< Diffusion, Viscosity, ThermalConductivity, CoeffType >::k(), Antioch::MixtureAveragedTransportEvaluator< Diffusion, Viscosity, ThermalConductivity, CoeffType >::mu(), Antioch::MixtureAveragedTransportEvaluator< Diffusion, Viscosity, ThermalConductivity, CoeffType >::mu_and_k(), Antioch::MixtureAveragedTransportEvaluator< Diffusion, Viscosity, ThermalConductivity, CoeffType >::mu_and_k_and_D(), Antioch::KineticsType< CoeffType, VectorCoeffType >::operator()(), Antioch::NASA7CurveFit< CoeffType >::s_over_R(), Antioch::NASA9CurveFit< CoeffType >::s_over_R(), set_zero(), tester(), vectester(), Antioch::SigmaBinConverter< VectorCoeffType >::y_on_custom_grid(), and zero_clone().
|
inline |
Definition at line 156 of file vector_utils.h.
|
inline |
Definition at line 157 of file vexcl_utils.h.
|
inline |
Definition at line 165 of file valarray_utils.h.
References zero_clone().
|
inline |
Definition at line 165 of file eigen_utils.h.
Referenced by set_zero(), and zero_clone().
|
inline |
|
inline |
Definition at line 167 of file vexcl_utils.h.
Referenced by eval_index().
|
inline |
Definition at line 176 of file valarray_utils.h.
References zero_clone().
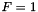 )
) 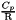
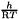 and
and 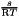 .
. ![\[ \log_{10}\left(F\right) = \frac{\log_{10}\left(F_{\text{cent}}\right)} {1 + \left[ \frac{\log_{10}\left(P_r\right) + c} {n - d \cdot \left[\log_{10}\left(P_r\right) + c\right]} \right]^2} \]](form_65.png)
![\[ \begin{split} P_r & = [\mathrm{M}] \frac{k_0}{k_\infty} \\ n & = 0.75 - 1.27 \log_{10}\left(F_{\text{cent}}\right) \\ c & = - 0.40 - 0.67 \log_{10}\left(F_{\text{cent}}\right) \\ d & = 0.14 \\ F_{\text{cent}} & = (1 - \alpha) \cdot \exp\left(-\frac{T}{T^{***}}\right) + \alpha \cdot \exp\left(-\frac{T}{T^*}\right) + \exp\left(-\frac{T^{**}}{T}\right) \end{split} \]](form_66.png)
![\[ \begin{split} \frac{\partial F_{\text{cent}}}{\partial T} & = \frac{\alpha - 1}{T^{***}} \cdot \exp\left(-\frac{T}{T^{***}}\right) - \frac{\alpha}{T^{*}} \cdot \exp\left(-\frac{T}{T^*}\right) + \frac{T^{**}}{T^{2}} \exp\left(-\frac{T^{**}}{T}\right) \\ \frac{\partial \log_{10}\left(F_\text{cent}\right)}{\partial T} & = \frac{1}{\ln(10) F_\text{cent}}\frac{\partial F_\text{cent}}{\partial T} \\ \frac{\partial n}{\partial T} & = - 1.27 \frac{\partial \log_{10}\left(F_\text{cent}\right)}{\partial T} \\ \frac{\partial c}{\partial T} & = - 0.67 \frac{\partial \log_{10}\left(F_\text{cent}\right)}{\partial T} \\\\ \frac{\partial P_r}{\partial T} & = P_r \left(\frac{\partial k_0}{\partial T} \frac{1}{k_0} - \frac{\partial k_\infty}{\partial T} \frac{1}{k_\infty} \right)\\ \frac{\partial \log_{10}(P_r)}{\partial T} & = \frac{1}{\ln(10) P_r} \frac{\partial P_r}{\partial T} \\\\ \frac{\partial \log_{10}(F)}{\partial T} & = \frac{\partial \log_{10}\left(F_\text{cent}\right)}{\partial T} \frac{1}{1 + \left[\frac{\log_{10}\left(P_r\right) + c} {n - d\left(\log_{10}\left(P_r\right) + c\right)}\right]^2} - \log_{10}\left(F_\text{cent}\right) 2\left[\frac{\log_{10}\left(P_r\right) + c}{n - d \left[\log_{10}\left(P_r\right) + c\right]}\right]^2 \left[\frac{\frac{\partial \log_{10}\left(P_r\right)}{\partial T} + \frac{\partial c}{\partial T}} {\log_{10}\left(P_r\right) + c} - \frac{\frac{\partial n}{\partial T} - d \left[\frac{\partial \log_{10}\left(P_r\right)}{\partial T} + \frac{\partial c}{\partial T}\right]} {n - d \left[\log_{10}\left(P_r\right) + c\right]} \right] \frac{1}{\left[1 + \left[\frac{\log_{10}\left(P_r\right) + c} {n - d\left(\log_{10}\left(P_r\right) + c\right)} \right]^2 \right]^2} \\ & = \log_{10}\left(F\right) \left[\frac{\partial \log_{10}\left(F_\text{cent}\right)}{\partial T} \frac{1}{F_\text{cent}} - 2\left[\frac{\log_{10}\left(P_r\right) + c}{n - d \left[\log_{10}\left(P_r\right) + c\right]}\right]^2 \left[\frac{\frac{\partial \log_{10}\left(P_r\right)}{\partial T} + \frac{\partial c}{\partial T}} {\log_{10}\left(P_r\right) + c} - \frac{\frac{\partial n}{\partial T} - d \left[\frac{\partial \log_{10}\left(P_r\right)}{\partial T} + \frac{\partial c}{\partial T}\right]} {n - d \left[\log_{10}\left(P_r\right) + c\right]} \right] \frac{1}{1 + \left[\frac{\log_{10}\left(P_r\right) + c} {n - d\left(\log_{10}\left(P_r\right) + c\right)} \right]^2} \right] \\ \frac{\partial F}{\partial T} & = \ln(10) F \frac{\partial \log_{10}\left(F\right)}{\partial T} \\\\\\\\ \frac{\partial P_r}{\partial c_i} & = \frac{k_0}{k_\infty} \\ \frac{\partial \log_{10}(P_r)}{\partial c_i} & = \frac{1}{\ln(10) P_r} \frac{\partial P_r}{\partial c_i} = \frac{1}{\ln(10) [\mathrm{M}]}\\\\ \frac{\partial \log_{10}\left(F\right)}{\partial c_i} & = -\frac{\log_{10}^2\left(F\right)}{\log_{10}\left(F_\text{cent}\right)} \frac{\partial \log_{10}\left(P_r\right)}{\partial c_i} \left(1 - \frac{1}{n - d\left[\log_{10}\left(P_r\right) + c\right]}\right) \left(\log_{10}\left(P_r\right) + c\right) \\ \frac{\partial F}{\partial c_i} & = \ln(10) F \frac{\partial \log_{10}\left(F\right)}{\partial c_i} \end{split} \]](form_67.png)